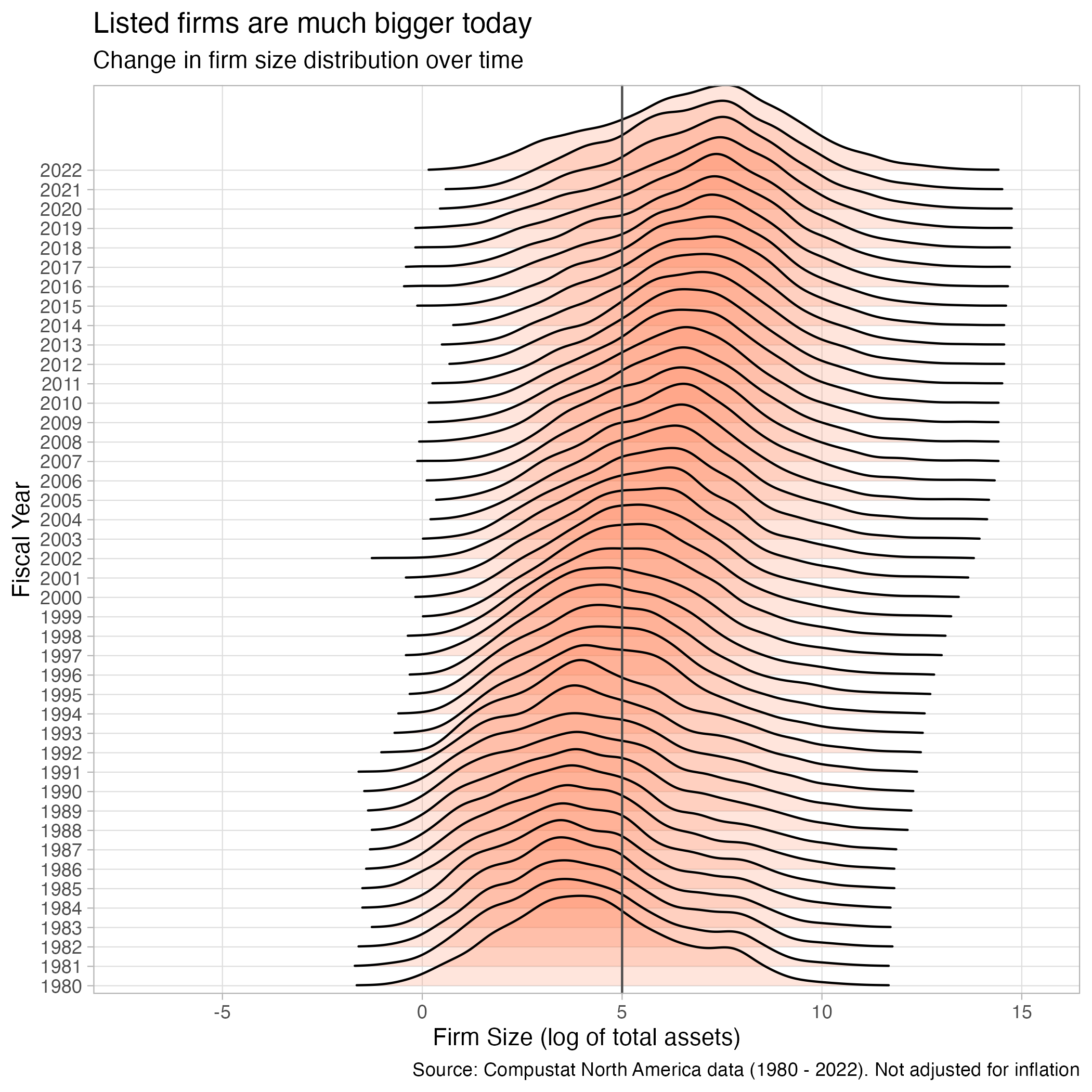
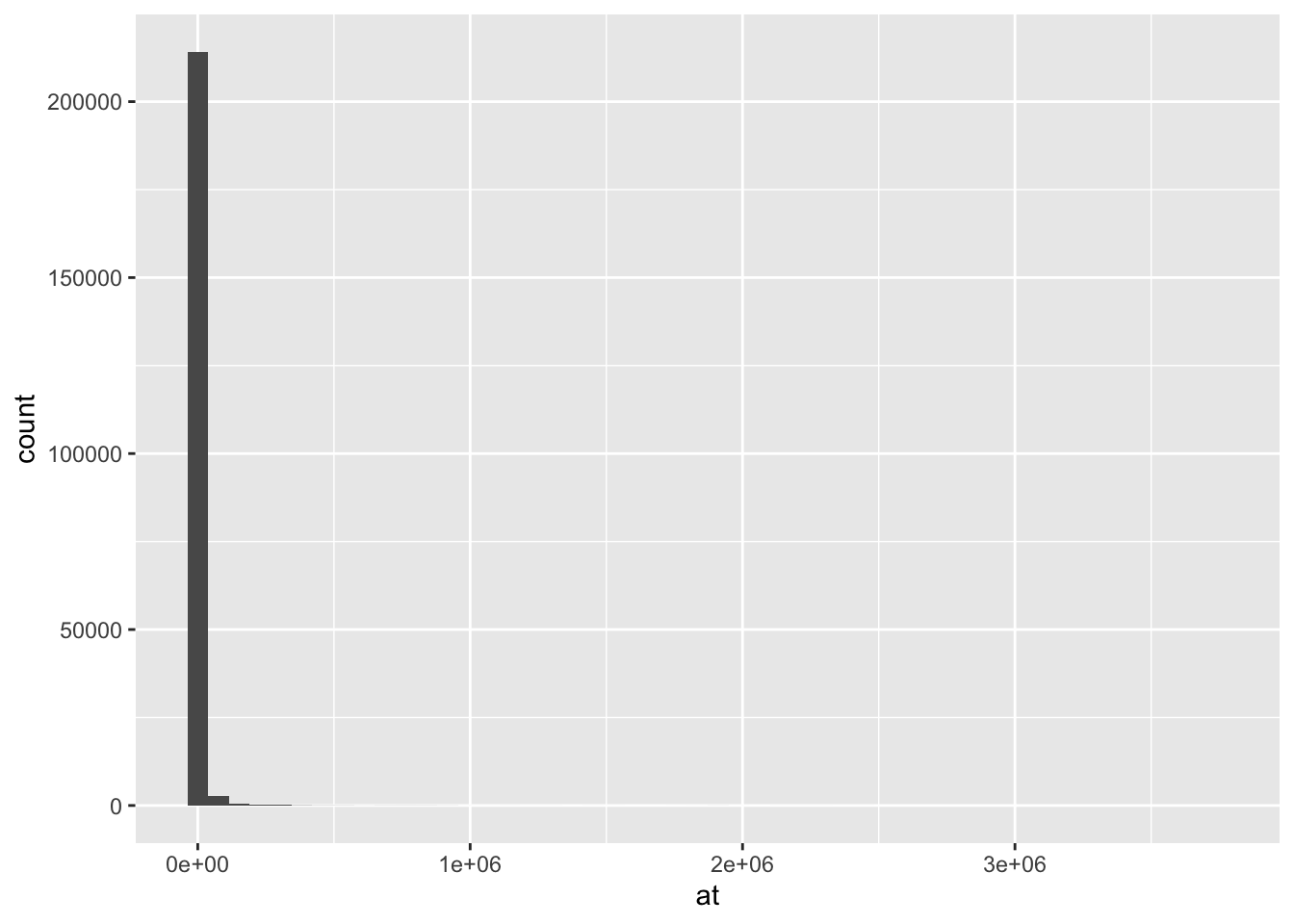
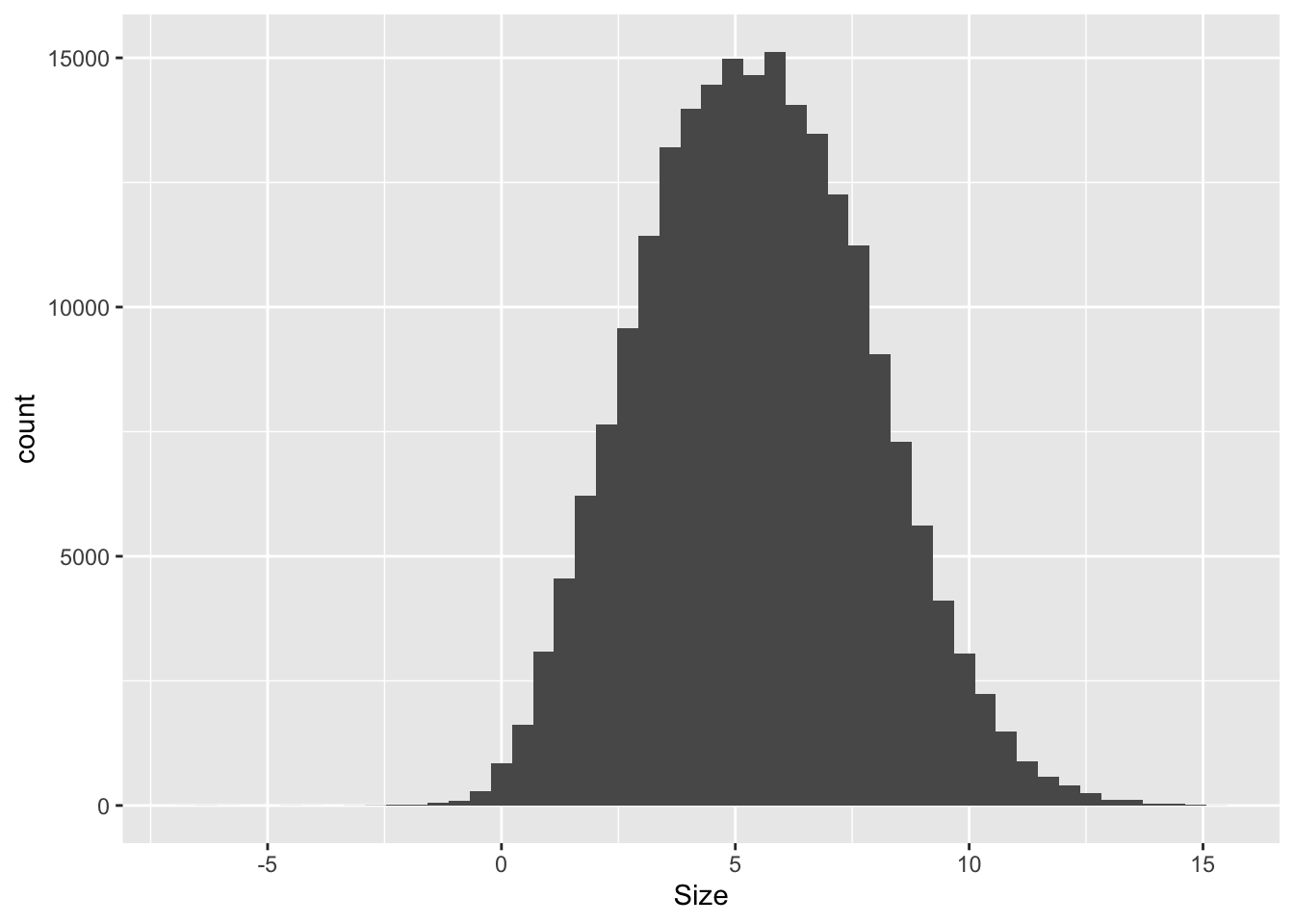
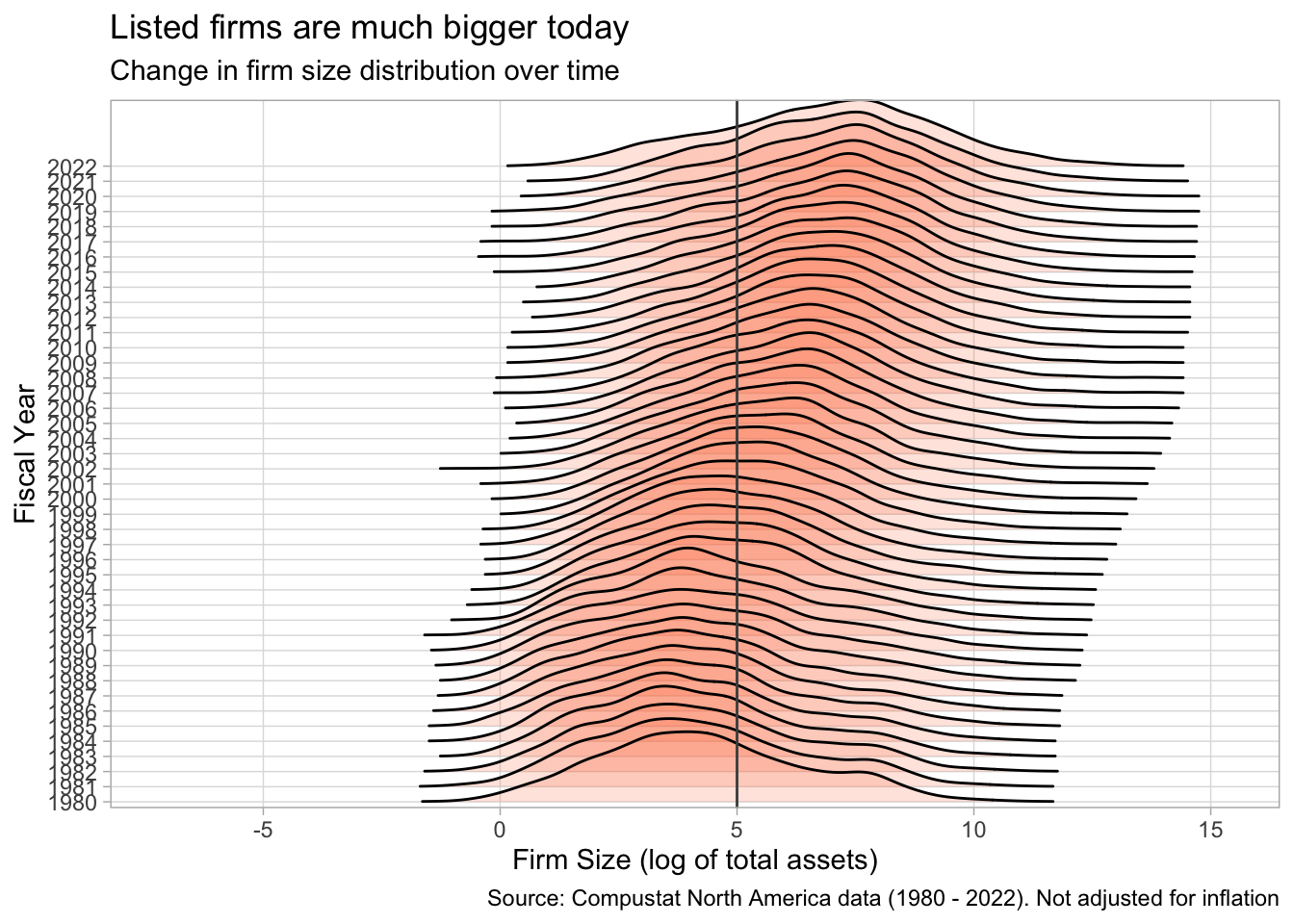
Rows: 271,143
Columns: 29
$ gvkey <chr> "001001", "001001", "001001", "001003", "001003", "001003", "…
$ linkprim <chr> "P", "P", "P", "C", "C", "C", "C", "C", "C", "P", "P", "P", "…
$ liid <chr> "01", "01", "01", "01", "01", "01", "01", "01", "01", "01", "…
$ linktype <chr> "LU", "LU", "LU", "LU", "LU", "LU", "LU", "LU", "LU", "LU", "…
$ permno <dbl> 10015, 10015, 10015, 10031, 10031, 10031, 10031, 10031, 10031…
$ datadate <date> 1983-12-31, 1984-12-31, 1985-12-31, 1983-12-31, 1984-12-31, …
$ fyear <dbl> 1983, 1984, 1985, 1983, 1984, 1985, 1986, 1987, 1988, 1980, 1…
$ indfmt <chr> "INDL", "INDL", "INDL", "INDL", "INDL", "INDL", "INDL", "INDL…
$ consol <chr> "C", "C", "C", "C", "C", "C", "C", "C", "C", "C", "C", "C", "…
$ popsrc <chr> "D", "D", "D", "D", "D", "D", "D", "D", "D", "D", "D", "D", "…
$ datafmt <chr> "STD", "STD", "STD", "STD", "STD", "STD", "STD", "STD", "STD"…
$ tic <chr> "AMFD.", "AMFD.", "AMFD.", "ANTQ", "ANTQ", "ANTQ", "ANTQ", "A…
$ cusip <chr> "000165100", "000165100", "000165100", "000354100", "00035410…
$ conm <chr> "A & M FOOD SERVICES INC", "A & M FOOD SERVICES INC", "A & M …
$ curcd <chr> "USD", "USD", "USD", "USD", "USD", "USD", "USD", "USD", "USD"…
$ fyr <dbl> 12, 12, 12, 12, 12, 1, 1, 1, 1, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5,…
$ at <dbl> 14.080, 16.267, 39.495, 8.529, 8.241, 13.990, 14.586, 16.042,…
$ ceq <dbl> 7.823, 8.962, 13.014, 6.095, 6.482, 6.665, 7.458, 7.643, -0.1…
$ che <dbl> 4.280, 1.986, 2.787, 2.023, 0.844, 0.005, 0.241, 0.475, 0.302…
$ dlc <dbl> 0.520, 0.597, 8.336, 0.250, 0.350, 0.018, 0.013, 0.030, 7.626…
$ dltt <dbl> 4.344, 4.181, 11.908, 0.950, 0.600, 4.682, 3.750, 5.478, 0.10…
$ ib <dbl> 1.135, 1.138, 2.576, 1.050, 0.387, 0.236, 0.793, -0.525, -7.8…
$ oiadp <dbl> 1.697, 1.892, 5.238, 2.089, 0.732, 0.658, 1.933, -0.405, -4.3…
$ sale <dbl> 25.395, 32.007, 53.798, 13.793, 13.829, 24.189, 36.308, 37.35…
$ exchg <dbl> 14, 14, 14, 19, 19, 19, 19, 19, 19, 11, 11, 11, 11, 11, 11, 1…
$ costat <chr> "I", "I", "I", "I", "I", "I", "I", "I", "I", "A", "A", "A", "…
$ fic <chr> "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA", "USA"…
$ priusa <chr> "01", "01", "01", "01", "01", "01", "01", "01", "01", "01", "…
$ sic <chr> "5812", "5812", "5812", "5712", "5712", "5712", "5712", "5712…